Microfabricated Fixed-Target Supports for Protein Crystallography
Over the last decade, brilliant, coherent femtosecond X-ray free-electron lasers (1012 photons/pulse) have revolutionized structural biology by enabling ultrafast, room-temperature protein structure and dynamics studies. Using a diffract-before-destroy approach, even weakly-diffracting micro or nanocrystals of hard-to-crystallize proteins can be studied, but challenges exist in efficiently delivering thousands of crystals to the X-ray beam while maintaining crystal integrity in a vacuum environment. Microfabricated fixed-target supports are an exciting alternative to widely used liquid jet-based technologies as they offer distinct advantages like significantly lower sample consumption, control over sample distribution, and the ability to incorporate stimuli like ligands, caged reactants, or electric fields “on-chip” for dynamic time-resolved experiments.
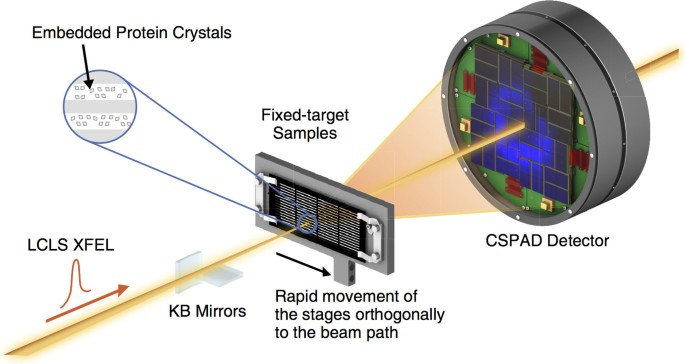
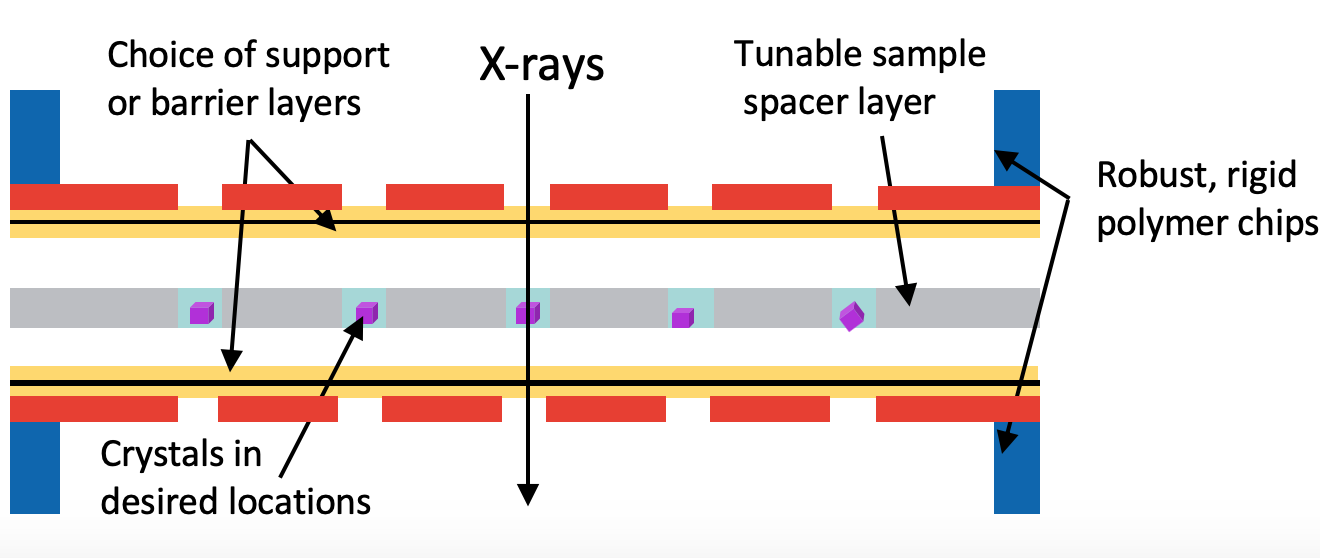

This work focuses on the development of robust, hot-embossed cyclic olefin copolymer (COC) fixed-target supports with ultra-thin, tunable water barrier films made from COC and graphene. These supports are inexpensive, maintain protein crystal hydration over extended periods of time, while contributing minimally to the X-ray scatter background compared to existing silicon or polymer-based supports. The enclosing COC thin films can be functionalized and patterned using UV-initiated photo-grafting of acrylic acid monomers, which can be further derivatized to create different charge or affinity binding moieties that act as “universal” protein crystal nucleation sites. This enables precise control of protein crystal sample distribution on fixed targets that can drastically improve sample hit-rates and reduce data collection time to a few minutes. Specific protein binding-driven crystal nucleation provides an alternative strategy for protein crystallization, particularly for proteins that are difficult to express and crystallize or have slow crystallization kinetics.